Project Introduction
The training of advanced deep learning algorithms for medical image interpretation requires precisely annotated datasets, which is laborious and expensive. Therefore, this research investigates state-of-the-art active learning methods for utilising limited annotations when performing automated left ventricle segmentation in echocardiography. Our experiments reveal that the performance of different sampling strategies varies between datasets from the same domain. Further, an optimised method for representativeness sampling is introduced, combining images from feature-based outliers to the most representative samples for label acquisition. The proposed method significantly outperforms the current literature and demonstrates convergence with minimal annotations. We demonstrate that careful selection of images can reduce the number of images needed to be annotated by up to 70%. This research can therefore present a cost-effective approach to handling datasets with limited expert annotations in echocardiography.
Fig. 1 A Sample image from the Unity dataset
Dataset
Two different datasets of cardiac imaging are used for our experiments:
the CAMUS dataset and the Unity dataset.
CAMUS dataset is a public, fully annotated dataset for 2D echocardiographic assessment that
contains 500 images from 2D apical four-champers and two-champers view sequences.
All the details of the CAMUS dataset are available on the official website (https://www.creatis.insa-lyon.fr/Challenge/camus/index.html).
We split CAMUS dataset into 70% (350 images) for training, 15% (50 images for validation), and 15% (50 images) for testing.
Unity datasetis a private dataset extracted from 1224 videos of the apical four-chamber echocardiographic view retrieved
from Imperial College Healthcare NHS Trust's echocardiogram database.
The images are obtained using ultrasound equipment from GE and Philips manufacturers.
The acquisition of these images is ethically approved by the Health Regulatory Agency
(Integrated Research Application System identifier 243023).
It contains 2800 images sampled from different points in the cardiac cycle and labelled by a pool of
experts using our in-house online labelling platform (https://unityimaging.net).
This dataset was used for model developments (i.e., training and validation) and split into 70% for training, 15% for validation and 15% for testing.
Network Architecture
The customised Bayesian U-Net architecture is illustrated in Fig. 2.
First, we used Conv2D with a kernel size of 3x3, Batch normalisation, and a "RELU" activation function for
the encoder blocks, followed by Maxpooling 2x2. Then, a Dropout layer is added with a Dropout probability
of 0.1.
At the centre, Bottleneck, two Conv2D followed by a Dropout layer with a Dropout probability of 0.25 are used.
Next, for the decoder blocks, we used Conv2DTranspose with a kernel size of 2x2, followed by a
concatenation layer, a Dropout layer with a Dropout probability of 0.1, Batch normalisation, and a
"RELU" activation. Then, two Conv2D with a kernel size of 3x3, Batch normalisation, and a "RELU"
activation. Finally, a Conv2D with a kernel size of 1x1 with "Sigmoid" activation is added for the
output layer.
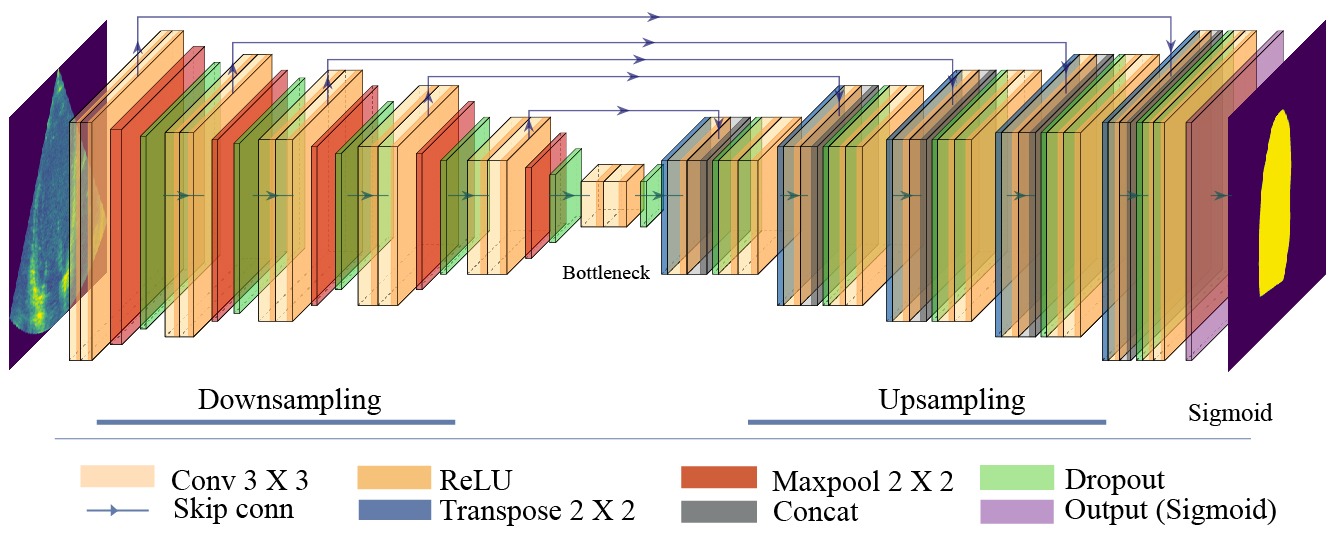
Fig. 2 The customised Bayesian U-Net architecture
Implementation
A standard Active Learning (AL) methodology is applied throughout, encompassing four steps: training the
U-Net model on the initial annotated data (L),
calculating the model uncertainty scores or representativeness scores on the unlabeled pool of data (U),
selecting the top-ranked batch of images (K) to obtain their labels from oracle and add them to L and remove
them from U, and finally retrain the model on the updated L.
These steps are repeated until the optimal number of AL iterations is reached.
Random sampling and a variety of different selective sampling approaches are used for
selecting the next batch of images from the unlabelled pool.
The most common AL selection strategy is uncertainty sampling, where the most uncertain unlabelled images are
quiried for annotation. Such uncertainty methods include: Classification Uncertainty (Pixel-wise), Predictive entropy,
and Monte Carlo droupout ensemble-based methods using entropy, variance (Var), variation ratio (Var_ratio),
standard diviation (STD), Coeffcient of variation (Coef_var),
and Bayesian active learning with disagreement (BALD).
Training settings: Tensorflow and Keras frameworks are used for the development of DL models, and training was conducted using an Nvidia RTX3090 GPU. U-Net was trained using binary cross-entropy loss and ADAM optimiser with a learning rate of 0.0001 for 200 epochs. Images were resized to 512x512, and a fixed batch size of 8 was applied.
For small datasets A and B, we selected 10% of the initial training data as L, and U will be the remaining 90%.
For the larger dataset, we chose 4% as the initial L, and the remaining is for the U, which will be
used as an oracle. Finally, to have a fair comparison between methods, we trained the model on the
initially labelled data of
the three datasets and used the same model's weights for training the following AL iterations.
Evaluation
We evaluated the model after every AL iteration using the Dice-Coefficient (DC) metric, and it is computed between the ground truth and the inferred prediction for each image in the testing dataset. Then the mean of Dice scores of all images is calculated to present the model's accuracy. Each AL selection strategy is trained three times, and the average of the DC at each AL iteration is computed and used to plot the results.
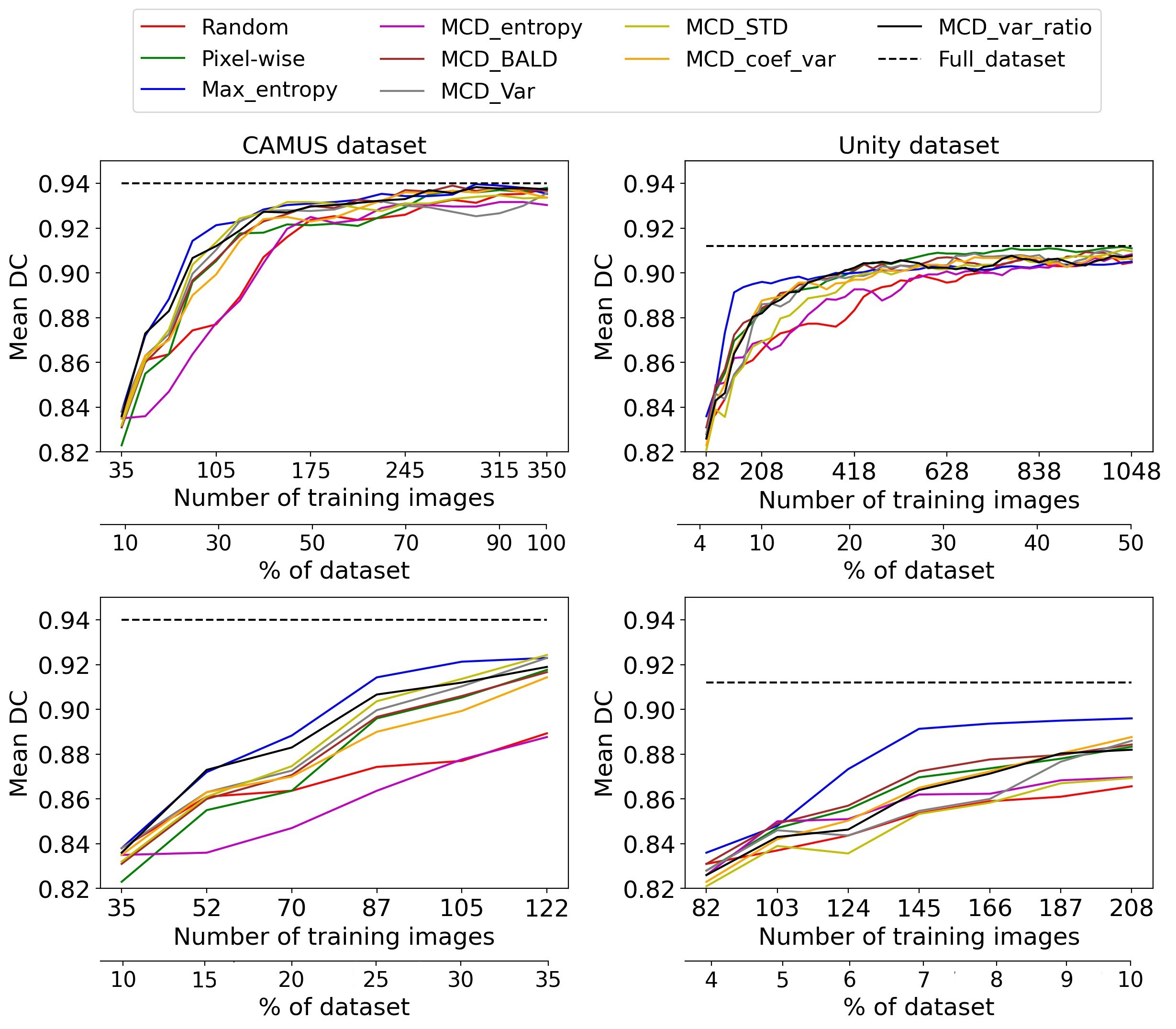
Fig. 3 Active learning performance on CAMUS and Unity datasets
